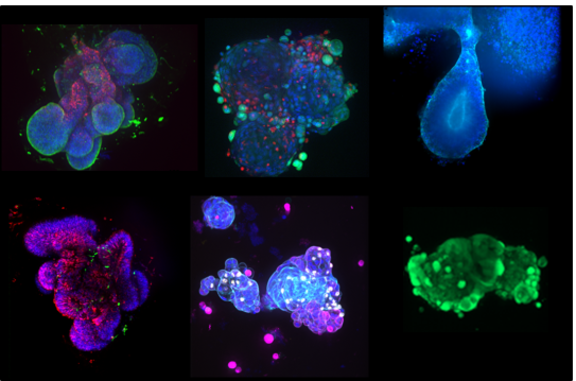
Patient-derived Microtumors
Exploring biology and application of patient-derived microtumors for drug development and personalized medicine
Current approaches to genomics-based personalized medicine have demonstrated the potential to improve the clinical course of cancer, particularly in patients with no hope of treatment beyond the current "one size fits all" approach. However, only a minority of patients currently experience clinical benefit. To improve this situation, it will be necessary to analyze the tumor disease of the individual patient on a broad basis, i.e. taking into account the tumor microenvironment as well as specific changes in the transcriptome, proteome, metabolome and microbiome. This allows more efficient stratification of individual patients compared to genome-based approaches, e.g. by identifying potential mono- and combination therapies beyond tumor mutations based on already clinically approved agents (off-label use) and identifying targets for novel therapies (including immunotherapies). Parallel testing of identified agents and their combinations in patient-derived ex vivo model systems allows quantification of clinical efficacy in clinically relevant time frames.
Projects
- SolidCarT- Modular mini-factories for autonomous production of CAR-T cells
- Patient-derived microtumors and autologous immune cells from brain tumor tissue for drug testing and precision oncology
- Wellcome LEAP HOPE- Organ-on-a-Chip Test Systems for the Development of Novel Cancer Therapies
- ELEVATE - Bioelectronic platform for personalized cancer therapy based on 3D-printed spheroids and patient-derived microtumors
- PRIMO- Personalized Medicine for Tailored Cancer Therapies






